Publications
Research articles
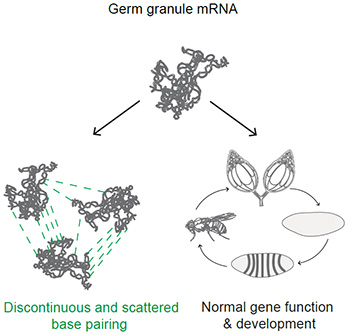
“We show that mRNAs remain well-folded upon localization to Drosophila germ granules wherein they organize into RNA clusters. This clustering is facilitated by base pairing among scattered and discontinuous stretches of bases appearing on the surface of folded RNAs, providing multivalency to clustering but exhibits low probability for sustained interactions. Notably, RNA structures with exposed GC-rich sequences enable sustained base pairing but also disrupt fly development. Thus, proper RNA folding safeguards mRNAs against detrimental effects of exposed GC-rich sequences.”
Tian, S., Nguyen, H., Ziqing, Y. Rouskin, S., Thirumalai, D., Trcek, T.
BioRxiv (2024); https://doi.org/10.1101/2024.05.31.596852
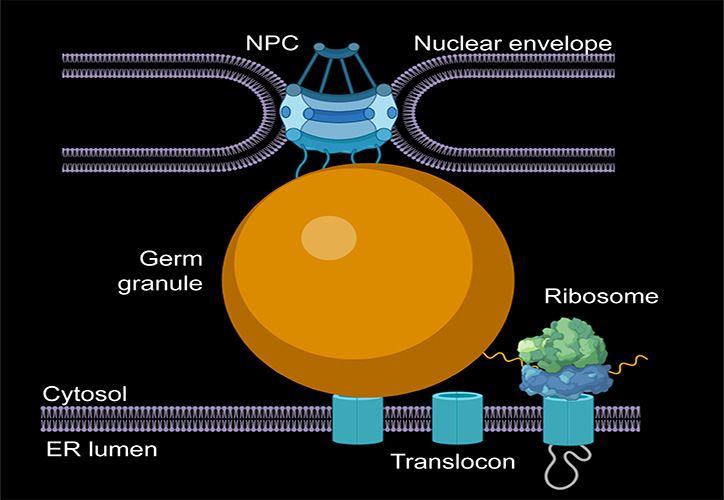
“We characterized the role of mRNAs and proteins in the formation of germ granules in Drosophila using super-resolution microscopy and genetic analysis. We found that proteins rather than mRNAs drive the formation of germ granules in Drosophila. Thus, the protein-driven formation of germ granules is mechanistically distinct from the RNA-dependent condensation observed for other RNA granules such as stress granules and P-bodies.”
Curnutte, H.A*, Lan, X.*, Sargen, M.*, Ao Ieong, S.M., Campbell, H.Kim, Y. Liao, D., Lazar, S.B., Trcek, T. (2023) “Cell Reports. 42, 112723”
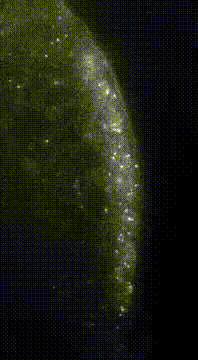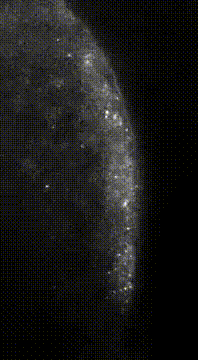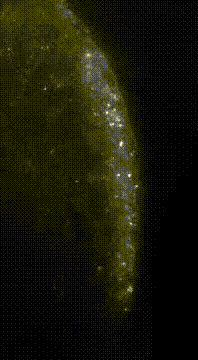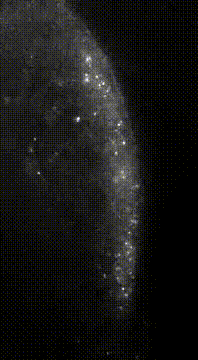
“Here we show that germ granule mRNAs self-assemble into homotypic clusters. The specificity for homotypic clustering is not generated by a particular RNA sequence and the interactions among clustered transcripts are transient. mRNA clustering is augmented under conditions that increase RNA concentration, such as upon enrichment in RNA granules, a process that appears
conserved in diverse cellular contexts and organisms”
“Sequence Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters.”
Tatjana Trcek*, Tyler E. Douglas, Markus Grosch, Yandong Yin, Whitby V.I. Eagle, Elizabeth R. Gavis, Hari Shroff, Eli Rothenberg, Ruth Lehmann* (2020); Mol Cell. 78(5)941-950
Kitchen Table Talk: Tatjana talks about mRNA self-assembly in germ granules
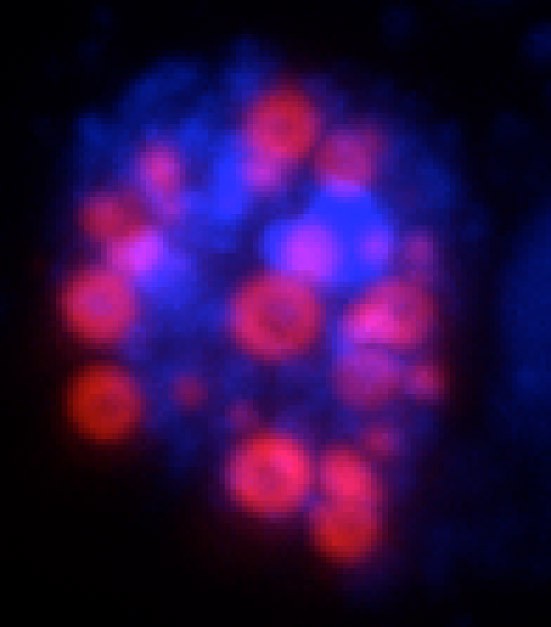
“The same scaffolding protein organizes cytoplasmic and nuclear granules that have similar biophysical properties but different composition and function relevant for germ cell biology. The cytoplasmic germ granules instruct the germ cell fate while the nuclear germ granules promote divisions of primordial germ cells.”
“Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells.”
Kistler, K.E.1, Trcek, T.*1, Hurd, R.T., Chen, R., Liang, F., Sall, J., Kato, M., Lehmann, R.* (2018) eLife https://doi.org/10.7554/eLife.37949
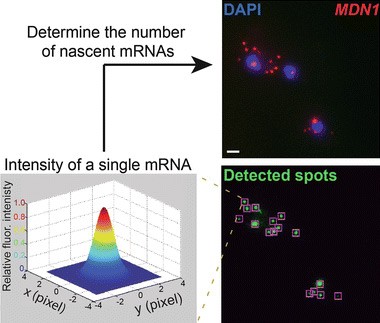
“Here we show how single-molecule fluorescent in situ hybridization (smFISH) can be used to measure mRNA decay of constitutively expressed genes in genetically and chemically unperturbed single budding yeast cells.”
“Measuring mRNA decay in budding yeast using single molecule FISH.”
Trcek, T*. Rahman, S., Zenklusen D.* (2018) Methods Mol Biol. 1720:35-54.
“We provide a detailed protocol of labeling of mRNAs with smFISH probes in Drosophila embryos and oocytes and a step-by-step protocol to detect and count single smFISH-labeled mRNAs in three dimensions in these Drosophila tissues.”
“mRNA quantification using single-molecule FISH in Drosophila embryos.”
Trcek, T*., Lionnet T., Shroff H., Lehmann R.* (2017). Nat Protocols. 12(7): 1326-1348
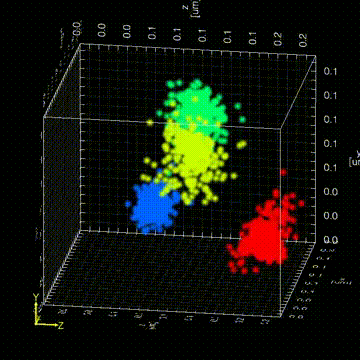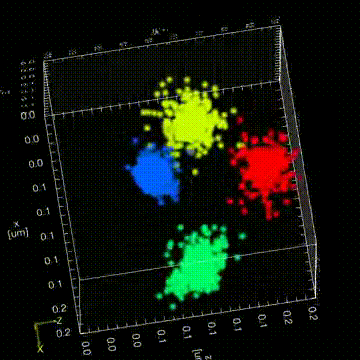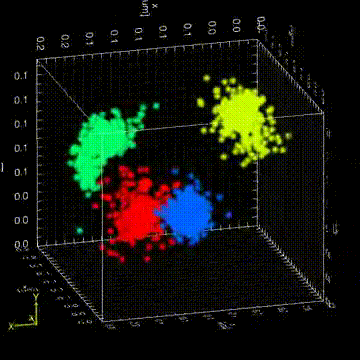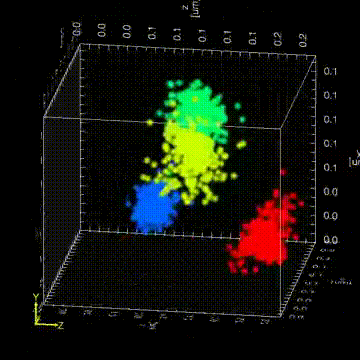
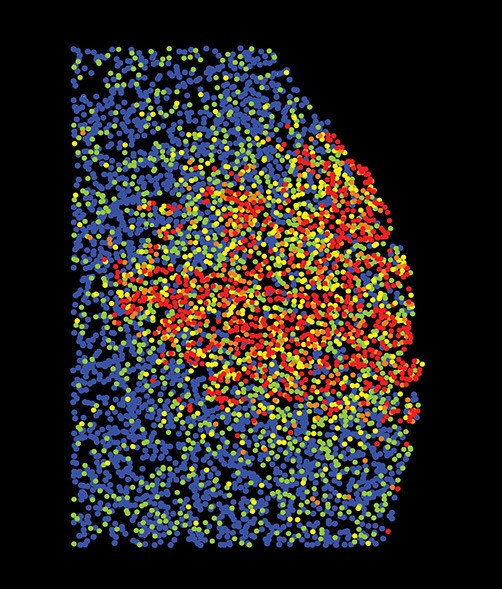
“The Drosophila germ granules accumulate a variety of mRNAs that instruct the germ cell fate. We show that mRNAs derived from the same gene form homotypic clusters, which occupy defined positions in the center or at the periphery of the granule while the core granule proteins that make the granules and recruit the mRNAs are homogeneously distributed within granules. This mRNA clustering has since been observed in a variety of RNA droplets indicating that it could be a conserved feature of phase separated condensates.”
“Drosophila germ granules are structured and contain homotypic mRNA clusters.”
Trcek, T., Grosch, M., York, A., Shroff, H., Lionnet, T., Lehmann, R. (2015) Nat Commun.6:7962 doi:10.1038/ncomms8962.
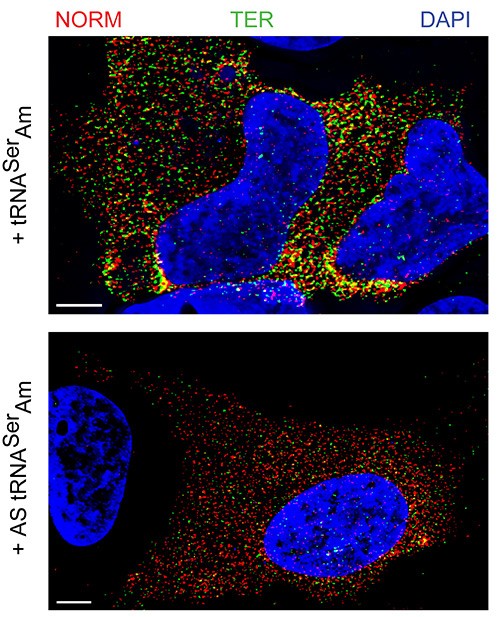
“In collaboration with Lynne E. Maquat, Rochester University, we showed that the mammalian Nonsense-Mediated Decay (NMD) occurs in the cytoplasm within the 400 nm zone surrounding the nuclear envelope. Thus, cells have evolved an efficient surveillance system that removes erroneous messages immediately upon exit from the nucleus.”
“Temporal and spatial characterization of nonsense-mediated mRNA decay.”
Trcek, T., Sato, H., Singer, R.H., Maquat, L.E. (2013). Genes Dev. 27(5): 541-51.
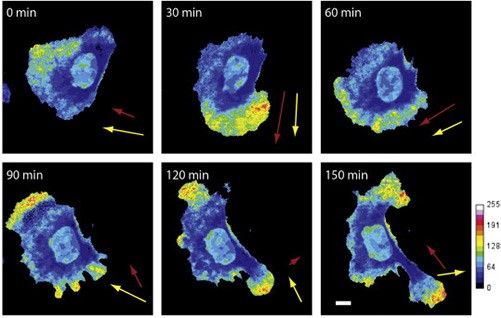
“Here, an analytical method is described that allows unbiased quantification of mRNA localization in fixed and live single cells.”
Park, H.Y., Trcek, T., Wells, A.L., Chao, J.A., Singer, R.H. (2012). Cell Reports. 1:179-184
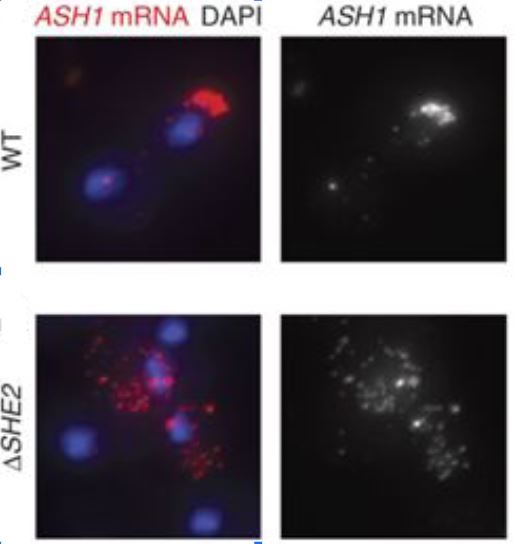
“We demonstrate how to perform fluorescent in situ hybridization (FISH) followed by single mRNA counting in genetically and chemically unperturbed single budding yeast cells.”
“Single mRNA counting using fluorescent in situ hybridization in budding yeast.”
Trcek, T., Chao, J.A., Larson, D.R., Park, H.Y., Zenklusen, D., Shenoy M.S., Singer, R.H. (2012). Nat Protocols. 7(2): 408-419
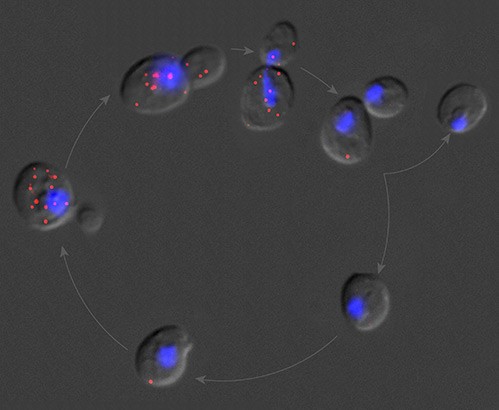
“Using smFISH coupled with mathematical modeling, we measured mRNA turnover in single yeast cells with minimal genetic or chemical perturbation. We discovered a promoter-regulated mRNA stability and demonstrated that cis mRNA sequences do not provide all the necessary information for post-transcriptional regulation.”
“Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast.”
Trcek, T., Larson, D.R., Moldon, A., Querry, C.C., Singer, R.H. (2011) Cell. 147(7): 1484-97.
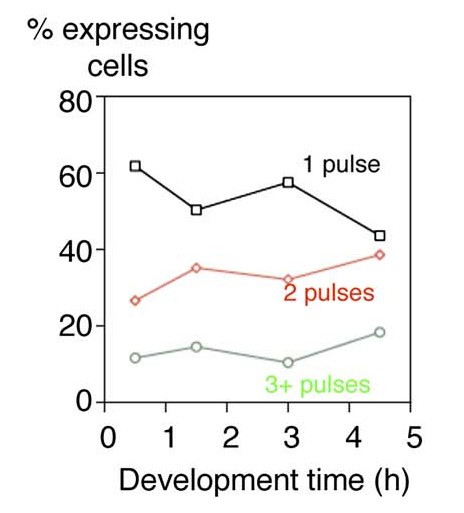
“In vivo mRNA imaging was used to visualize transcription of an endogenous developmental gene. Transcription was occurring in discrete transcriptional pulses at irregular intervals. This transcriptional discontinuity may allow a cell with a greater flexibility in making decisions during its gene-expression process.”
“Transcriptional pulsing of a developmental gene.”
Chubb, J. R., Trcek T., Shenoy, S.M., Singer, R.H. (2006). Curr Biol. 16(10): 1018-25.
“This study shows that the motility and targeting of peripherin mRNPs as well as their translational control and the assembly of an intermediate filament cytoskeletal system are linked together in a dynamic cotranslation.”
“Assembling an intermediate filament network by dynamic cotranslation.”
Chang, L., Y. Shav-Tal, Trcek, T., Singer, R.H., Goldman, R.D. (2006). J Cell Biol. 172(5): 747-58.
*Co-correspondence
1 Co-first authorship
